Denoising the Complex with AlphaFold 3
In this series: From AlphaFold2 to Boltz-2: The Protein Prediction Revolution
- Part 1: Deconstructing the Fold with AlphaFold2
- Part 2: Denoising the Complex with AlphaFold 3 (You are here)
- Part 3: From Fold to Function: Inside Boltz
- Part 4: Appendix (From Alphafold 2 to Boltz-2)
Introduction: The Generative Leap Beyond Single Proteins
In the summer of 2023, scientists spotted a worrying change in the COVID-19 virus: a new version carried a small but important mutation (E166V) in one of its key proteins, called the main protease. This mattered because our main antiviral pill, Paxlovid, works by fitting tightly into a pocket on that same protein, blocking the virus from multiplying. Researchers quickly fed the mutant sequence into AlphaFold 2[1] (the revolutionary model we deconstructed in Part 1[2]), and within minutes had an accurate 3D model showing the protein’s new shape.
But here’s the critical limitation: AlphaFold 2 could show how the protein itself changed, but not whether Paxlovid would still fit properly into its pocket. Predicting how a mutation affects a drug’s binding isn’t just about protein shape—it’s about modeling the detailed interaction between protein and drug. AlphaFold 2 was excellent at folding single proteins but wasn’t built to show how small molecules dock into altered pockets. Closing such gaps is exactly what motivated the development of AlphaFold 3[3][4].
To understand the leap required to solve this, it’s worth a quick look back at the brilliant but specialized engine of AlphaFold 2.
🧠 Refresher: Inside the AlphaFold 2 Engine
Evoformer & The Dialogue:
This module orchestrated a deep, two-way dialogue between evolutionary data (the MSA) and a geometric blueprint (the Pair Representation). It used an
Outer Product Mean to turn evolutionary signals into geometry, and an Attention Bias to let the geometry guide the search for more signals.
Triangular Attention:
This was the Evoformer's core reasoning engine. It enforced global 3D logic by ensuring that all residue-pair relationships on the blueprint were geometrically consistent with each other, allowing local information to rapidly influence the entire structure.
Structure Module & IPA:
After the blueprint was finalized, this deterministic module took over. Using a physically-inspired mechanism called
Invariant Point Attention (IPA), it translated the 2D map into a precise 3D atomic structure by operating in the local coordinate system of each residue.
AlphaFold 3 marks a significant departure from earlier designs, shifting the objective from predicting static protein conformations to modeling extensive, interactive molecular systems[5]. This transition represents not merely an incremental upgrade, but rather a profound shift in both philosophy and computational architecture, driven by two major developments:
- Transitioning from protein-specific to universal modeling: It adopts a universal framework capable of simultaneously processing a diverse set of molecular inputs.
- Adopting generative modeling over deterministic prediction: It employs a diffusion-based generative model that iteratively “denoises” a random cloud of atoms into a coherent, physically plausible structure.
In this post, we will thoroughly explore AlphaFold 3’s design philosophy and architecture. We will detail the technical choices enabling its universality, including its input handling, representational machinery, generative core, and training methodologies. Ultimately, this analysis aims to provide a clear, technical understanding of how AlphaFold 3 functions and significantly advances the frontiers of computational biology.
While AlphaFold 2 masterfully solved the 3D structure of individual proteins, it saw the "parts" but not the "machine." Real biological function—from how medicines work to how our genes are read—happens when these parts interact. AlphaFold 3 was completely redesigned to model this molecular dance, tackling the structure of entire complexes of proteins, DNA, RNA, and drug-like ligands all at once.
To achieve this, it made two profound shifts. First, it learned a universal language, representing standard protein and nucleic acid residues as single "tokens" while treating every atom of a small molecule as its own token. This flexibility allows it to model virtually any biological assembly. Second, it moved from a deterministic builder to a generative artist.
At its heart, the new Pairformer engine spends 48 cycles of intense computation to create a detailed geometric blueprint for the entire system. It listens only briefly to evolutionary hints (MSAs) before focusing on the fundamental geometry, figuring out how every token should be positioned relative to every other. This final blueprint is then handed to the diffusion module, which starts with a completely random "cloud" of atoms and, guided by the blueprint's thousands of constraints, patiently refines their positions. In a process of "denoising," it brings order from chaos, sculpting the fuzzy atomic cloud into a sharp, physically realistic 3D structure.
This generative approach has yielded huge gains, providing state-of-the-art accuracy for predicting how potential drugs bind to their protein targets. While challenges remain for very large molecular machines and complex RNA, and access is currently limited to a web server, AlphaFold 3 represents a fundamental paradigm shift—moving computational biology from predicting static puzzle pieces to modeling the dynamic, interacting machinery of life.
The Architectural Blueprint: From Atoms to Tokens
In Part 1, we saw how AlphaFold 2’s journey began with a protein’s 1D amino acid sequence. Its entire input system was expertly designed to process proteins, building a rich Multiple Sequence Alignment (MSA) and a geometric Pair Representation. This was a masterpiece for the world of proteins.
But what happens when you introduce a drug molecule, a strand of DNA, or an ion? These molecules have no “residues” in the traditional sense and no evolutionary cousins to build an MSA from. AlphaFold 2’s language was simply too specialized.
AlphaFold 3 solves this with a completely redesigned front end. It creates a universal language by shifting its focus from protein-specific residues to a more fundamental concept: tokens. This hybrid approach is the key to its versatility:
- For Proteins and Nucleic Acids: These molecules are made of repeating, standard units. AlphaFold 3 treats each amino acid (for proteins) or nucleotide (for DNA/RNA) as a single token, just as AlphaFold 2 viewed a protein. The geometry for each residue is an idealized template with perfect bond lengths and angles, retrieved from an internal library of known building blocks
- For Ligands and Other Molecules: Small molecules, drugs, and ions don’t have standard repeating units. Think of these molecules as keys designed to fit into the lock of a larger protein. In biology, these keys are called ligands. To handle this diversity, AlphaFold 3 adopts a more granular approach: every single heavy atom of a ligand becomes its own individual token. The geometry is a reference conformer generated on-the-fly from its 2D chemical structure using a tool like RDKit.[6]
To prepare these diverse inputs for its main engine, the model uses a new, more foundational two-stage pipeline.
Stage 1: The Atom-Level Foundation
Instead of jumping straight to a residue-level view, AlphaFold 3 first thinks about the system at the most basic level—the atoms. It gathers features for every single atom in the complex, creating initial atom-level single (c) and pair (p) representations. This information is then processed by a small transformer called the Input Embedder.
This is the first key innovation. The embedder uses Attention with Pair Bias—a mechanism we first saw in AlphaFold 2’s Evoformer when the geometric blueprint guided MSA attention—to allow the initial 3D distances between atoms to influence how they communicate. This “warm-up” cycle enriches the atom features with their immediate chemical and spatial context.
Stage 2: Assembling the Token-Level Representations
Once the atom-level features are refined, the model is ready to build the final blueprint for its main engine.
-
First, it creates the token-level single representation, s, by aggregating the feature vectors of the atoms that make up each token. For an amino acid, this means averaging the features of all its constituent atoms; for a ligand atom token, this step is trivial as it’s a 1-to-1 mapping.
-
Then, AlphaFold 3 creates the final token-level pair representation, z, by first pooling the underlying atom-pair data. It then enhances this foundation using the ‘outer sum’ and relative positional encoding technique borrowed from its predecessor.
At the end of this process, the model has successfully translated a chemically diverse biological scene into the clean, structured s and z tensors. This universal blueprint, now in a language its core engine can understand, is ready to be passed to the Pairformer.
The Representation Engine: Pairformer and Friends
With the universal s and z representations prepared, AlphaFold 3’s main reasoning engine takes over. Its job is to convert these simple initial inputs into a rich, coherent geometric blueprint for the entire molecular complex. This is where the model figures out all the critical interactions—how a drug fits into its pocket, or how two strands of DNA twist around each other.
This stage marks a major philosophical shift from AlphaFold 2, centered on a new communication strategy.
A New Communication Strategy: Dethroning the MSA
This is a major departure from AlphaFold 2’s design. Here’s a simple breakdown:
The Old Way (AlphaFold 2’s Evoformer): Imagine a 48-round debate between two experts: an “Evolution Expert” (using the MSA) and a “Geometry Expert” (using the pair representation). In every round, they exchanged notes, with the MSA playing a constant, essential role.
The New Way (AlphaFold 3’s Pairformer): The “Evolution Expert” gives a quick, 5-minute briefing at the very beginning. This information is passed to the “Geometry Expert” (the pair representation), and then the Evolution Expert leaves the room. For the remaining 48 cycles, the Geometry Expert works almost entirely alone, only communicating with the individual token features.
Why the change? AlphaFold 3 needs to model molecules that have no evolutionary history. By sidelining the MSA, the model is forced to rely less on evolutionary patterns and more on the fundamental, universal rules of chemistry and physics.
The Supporting Acts: A Quick Boost from Old Friends
Before the main loop begins, AlphaFold 3 gets a quick head start from external data, using tricks that will be very familiar to readers of Part 1.
-
Template Module: Just as in AlphaFold 2, if known structures of similar molecules exist, their geometric data is refined and added as a bias to the main pair representation (z), providing a powerful initial guess.
-
MSA Module: The MSA is now processed by only a few lightweight blocks and then discarded. The information is extracted using the same core mechanisms from the Evoformer: an Outer Product Mean operation summarizes co-evolutionary signals to update z, and Row-wise Gated Self-Attention allows the geometry in z to briefly bias the MSA attention.
Main Act: The Pairformer
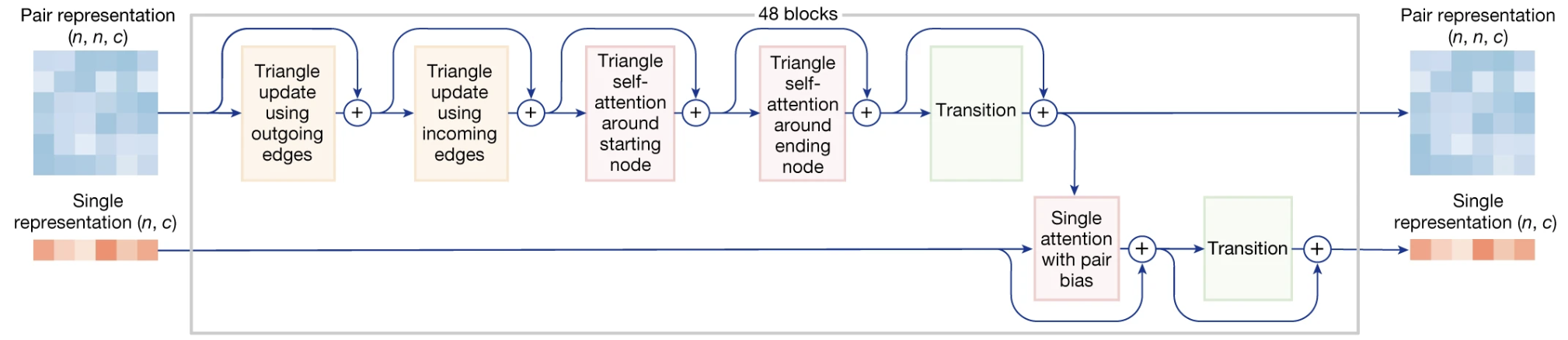 Figure 1. A detailed look inside a single block of the AlphaFold 3 Pairformer. The top pathway refines the geometric blueprint (the pair representation, $z$) using four triangle operations. The bottom pathway updates the token features (the single representation, $s$). Information flows from the refined pair representation to the single representation via ‘Single attention with pair bias’, allowing the geometric blueprint to guide the understanding of each token. This block is repeated 48 times.
Figure 1. A detailed look inside a single block of the AlphaFold 3 Pairformer. The top pathway refines the geometric blueprint (the pair representation, $z$) using four triangle operations. The bottom pathway updates the token features (the single representation, $s$). Information flows from the refined pair representation to the single representation via ‘Single attention with pair bias’, allowing the geometric blueprint to guide the understanding of each token. This block is repeated 48 times.
With the hints from templates and MSAs integrated, the Pairformer begins its 48-cycle refinement loop[7]. While it replaces the Evoformer, its power comes from the exact same core geometric reasoning engine that made AlphaFold 2 so successful.
The core logic is identical to AlphaFold 2’s, designed to enforce global geometric consistency. The model reasons about triplets of tokens (i, j, k) to ensure the map of their relationships is physically plausible. It uses two key mechanisms refined from the original Evoformer:
-
Triangle Updates: These operations perform a fast, “brute-force” update by combining information from all possible triangles, strengthening local geometric signals across the entire map.
-
Triangle Attention: This is a more sophisticated operation where the relationship between two points (i, j) selectively attends to information from other pairs. This attention is guided by a learned bias from the third edge of the triangle, allowing the model to propagate critical information over long distances and create a globally coherent structure. This remains the primary reasoning engine of the model.
Think of the pair representation (z) as the global “blueprint” detailing all relationships, while the single representation (s) is a list of “properties” for each token. Within a single Pairformer block, the triangle operations first exclusively refine the global blueprint z. Then, in a strictly one-way communication step, Single Attention with Pair Bias uses this improved blueprint to update the properties of each individual token in s. Critically, there is no reverse channel for the token properties to influence the blueprint within the same block.
This one-way information flow within the main loop (from z to s) is repeated 48 times, producing the final, rich blueprint —composed of the final z to s representations— that is handed off to the diffusion module.
The Generative Core: Sculpting Structures from Chaos 🎨
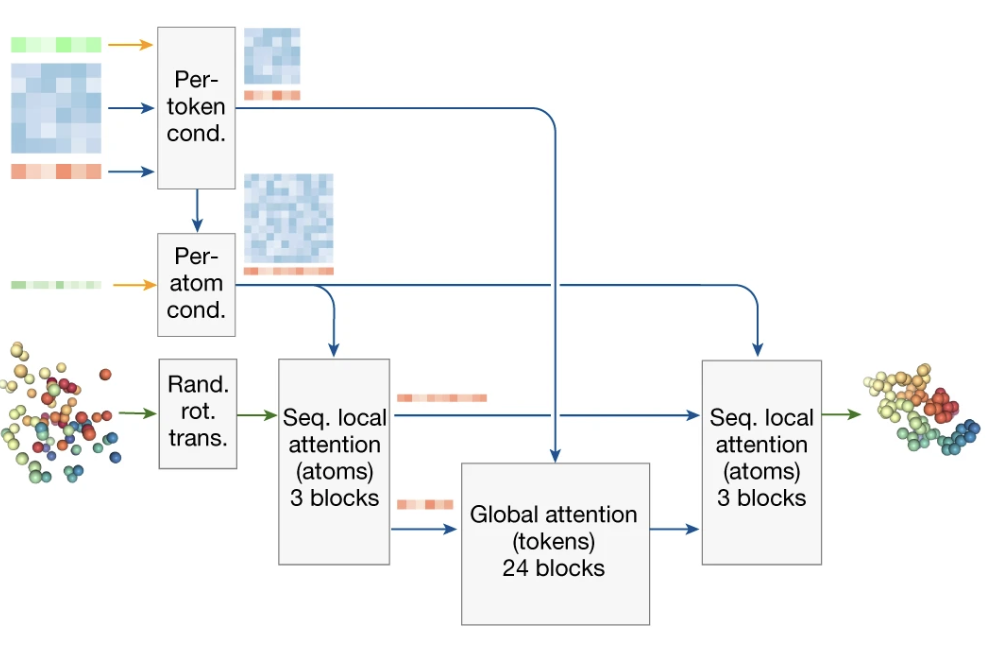 Figure 2. The architecture of the AlphaFold 3 Diffusion Module. At each denoising step, the module takes the current noisy atom coordinates (left) and the conditioning ‘blueprint’ from the Pairformer (top-left) as inputs. It processes this information in three main stages: a block of local attention over atoms, a large block of global attention over tokens, and a final block of local atom attention to make fine-grained adjustments. This coarse-to-fine process, guided by the conditioning blueprint at every stage, allows the model to produce a chemically realistic and globally coherent structure.
Figure 2. The architecture of the AlphaFold 3 Diffusion Module. At each denoising step, the module takes the current noisy atom coordinates (left) and the conditioning ‘blueprint’ from the Pairformer (top-left) as inputs. It processes this information in three main stages: a block of local attention over atoms, a large block of global attention over tokens, and a final block of local atom attention to make fine-grained adjustments. This coarse-to-fine process, guided by the conditioning blueprint at every stage, allows the model to produce a chemically realistic and globally coherent structure.
Here we arrive at the most radical departure from AlphaFold 2. The deterministic, Lego-like Structure Module is gone. In its place stands a generative AI model that operates on a completely different philosophy. Instead of building a structure piece by piece, AlphaFold 3 starts with a random, meaningless cloud of atoms and, like a master sculptor, patiently “carves” away the noise to reveal the final, coherent structure hidden within.
This sculptor, however, isn’t working from memory; it’s meticulously following a set of hyper-detailed instructions.
The Blueprint: Conditional Diffusion
The technical name for this process is a conditional diffusion model. While the mathematics are deep (see Appendix), the concept is beautifully elegant.
-
Learning by “Dissolving”: During training, the model learns the art of creation by mastering destruction. It takes thousands of correct, finished structures and observes them as they are systematically “dissolved” into a random fog of atoms by adding noise in many small steps. By watching this movie in reverse, it learns the precise path from order to chaos.
-
Sculpting by Denoising: At prediction time, the process is inverted. The model starts with a completely random cloud of atoms and, using its learned knowledge, reverses the dissolution step-by-step. Each step is a “denoising” operation—a single, confident chisel stroke that brings the chaotic cloud slightly closer to a chemically plausible structure.
The critical word here is conditional. The model is guided at every single step by the final blueprint—the s and z representations—generated by the Pairformer. This blueprint provides a powerful set of guiding constraints, defining which interactions are favorable (low-energy valleys) and which are forbidden (high-energy hills) to ensure the final sculpture is the one specified by the input [8].
Inside the Sculptor’s Mind: The Diffusion Transformer
Each denoising step, each “chisel stroke,” is not a simple operation. It’s a full-fledged computational process executed by another powerful Transformer, specifically designed to interpret and refine a noisy 3D scene. Its singular goal at each step is to look at the current jumbled atomic cloud and predict the exact noise vector that was added to corrupt the clean structure.
To do this, the Diffusion Transformer uses three “senses” to perceive the scene and the goal:
- The Noisy Scene (
xₜ): The current, jumbled 3D coordinates of all atoms in the system. - The Guiding Blueprint (
z_trunk): The final, incredibly rich pair representation from the Pairformer. This is held constant and acts as the unchanging “master plan” throughout the entire diffusion process. - A Sense of Time (
t): A learned time embedding. This is crucial because it tells the network how much noise to expect. At the beginning of the process (tis large), it knows the scene is very chaotic and its “chisel strokes” can be broad. Near the end (tis small), it knows the structure is almost finished and its adjustments must be subtle and precise.
A Denoising Step: A Three-Act Play
The transformer’s workflow for predicting the noise can be thought of as a three-act play, moving from a coarse perception to fine-grained action.
Act I: Perception (From Atoms to Tokens)
First, the sculptor gets a feel for the material. The transformer can’t immediately reason about the entire cloud of thousands of atoms. It first aggregates information from the noisy atom coordinates (xₜ) up to the token level. This creates a summarized feature vector for each protein residue, nucleotide, or ligand, effectively asking: “Given the current mess, what is the state of this particular piece?” This gives the model a manageable, high-level summary of the entire chaotic scene.
Act II: Global Strategy (Token-Level Attention with Blueprint Guidance)
This is the heart of the reasoning process, where the sculptor steps back, blueprint in hand, to plan the major moves. The transformer performs self-attention across all tokens. This is where the blueprint’s guidance is most critical. For any two tokens i and j, the attention score is calculated by combining two questions:
- “How do we relate right now?” This is a standard query-key dot product, assessing the relationship based on the tokens’ current, noisy state.
- “How should we relate in the final structure?” This is a powerful bias added directly to the score, which comes straight from the Pairformer’s blueprint:
z_trunk.
The full calculation is:
AttentionScore(i, j) = Query_i · Key_j + Bias(z_trunk_ij)
And yes, this is the exact same self-attention with pair bias we have been seeing time and time again! It is the core mechanism that powered AlphaFold 2’s Evoformer and even appeared in AlphaFold 3’s own Input Embedder. Its reappearance here underscores how fundamental this technique is for letting a geometric blueprint guide the attention process.
If the blueprint (z_trunk) says two tokens must be in close contact, the bias term will be huge, forcing the transformer to pay close attention to their relationship, regardless of how far apart they are in the current noisy cloud. This ensures the model’s global strategy is always aligned with the final goal.
Act III: Precise Execution (Atom-Level Refinement)
With a global strategy decided, the sculptor leans in to make the cuts. The new, globally-aware information for each token is broadcast back down to every single atom that belongs to it.
Armed with this rich context, a final set of attention layers operates directly at the atom level. These layers now have the full picture: they know their own noisy position, the state of the whole system, and the target geometry from the blueprint. This allows the model to make incredibly precise and coordinated predictions. It’s no longer just jiggling atoms; it’s nudging an oxygen on a ligand and a nitrogen on a protein in a concerted way that satisfies the hydrogen bond demanded by the global plan.
The final output of this three-act play is the transformer’s best guess for the noise vector (ε_θ). Subtracting this predicted noise from the current coordinates (xₜ - ε_θ) is the physical “chisel stroke”—a single, confident step on the long path from chaos back to a perfectly formed structure.
Training a Universal Molecular Modeler 🏋️
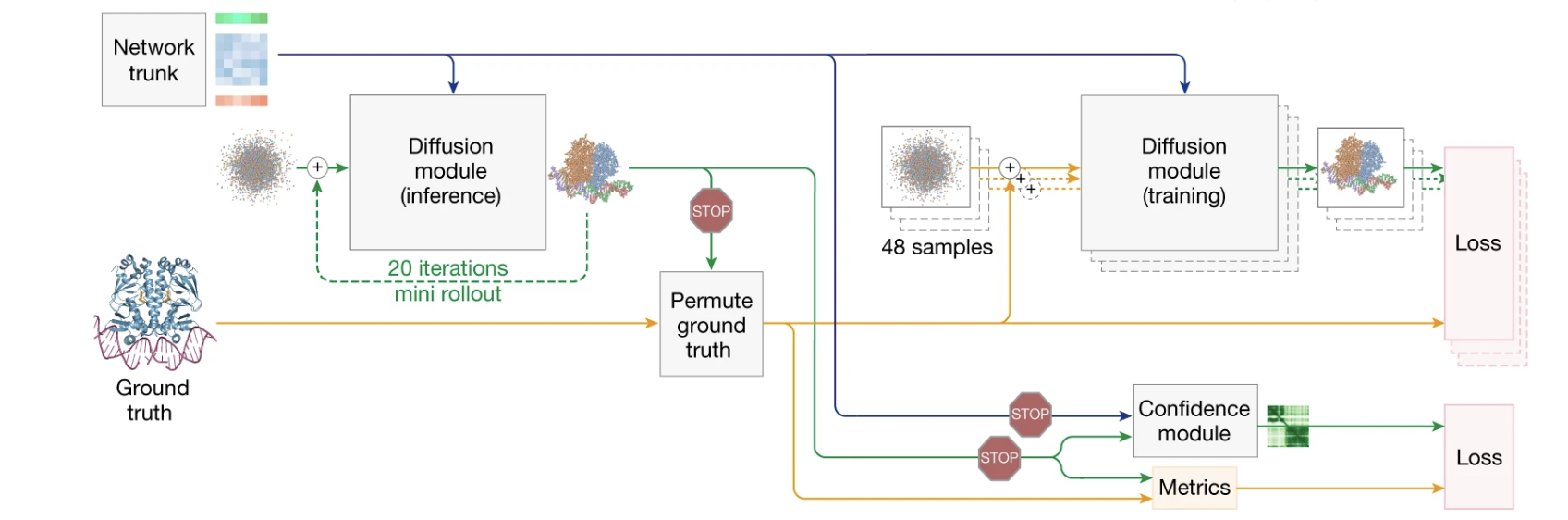 Figure 3. High-level overview of the AlphaFold 3 training setup. The diagram shows two parallel processes. The main training loop (right) teaches the Diffusion module to denoise a ground truth structure, guided by the network trunk’s output, to compute the main diffusion loss. A separate path (left) trains the Confidence module: a ‘mini rollout’ generates a sample structure, which is fed to the confidence module to predict its own accuracy (pLDDT, PAE, etc.). The stop-gradient symbols indicate that the confidence loss only updates the confidence module, not the main structure prediction network.
Figure 3. High-level overview of the AlphaFold 3 training setup. The diagram shows two parallel processes. The main training loop (right) teaches the Diffusion module to denoise a ground truth structure, guided by the network trunk’s output, to compute the main diffusion loss. A separate path (left) trains the Confidence module: a ‘mini rollout’ generates a sample structure, which is fed to the confidence module to predict its own accuracy (pLDDT, PAE, etc.). The stop-gradient symbols indicate that the confidence loss only updates the confidence module, not the main structure prediction network.
A revolutionary architecture is only half the story. To learn its complex task, AlphaFold 3 relies on an equally sophisticated training process. The goal isn’t just to teach the model to build accurate structures, but also to make it a reliable self-critic, aware of its own confidence. This is achieved by calculating two separate types of losses in parallel.
The Main Diffusion Loss
The main training loop teaches the diffusion module its core task: predicting the noise. For each training example, the model takes the ground truth structure, adds a random amount of noise, and then tries to predict the exact noise vector that was added. The difference between the model’s predicted noise and the true noise forms the primary diffusion loss.
This is supplemented by an auxiliary distogram loss on the Pairformer’s output, ensuring the model’s internal geometric blueprint is accurate even before the diffusion process begins.
The Confidence Loss via “Mini Rollout”
A brilliant model that doesn’t know when it’s wrong is dangerous. To make AlphaFold 3 a trustworthy tool, it’s also trained to predict its own accuracy. During training only, because running the full diffusion process at every step would be computationally expensive, the model uses a quick, 20-step ‘mini-rollout’ as a practical shortcut to generate approximate sample structures for the confidence head. At inference time, however, AlphaFold 3 always runs the complete diffusion rollout, providing the final, high-quality structure directly to the confidence module.
This generated structure is then compared to the ground truth to calculate the “real” error metrics (pLDDT, PAE, etc.). A separate Confidence module is then trained to predict these same error metrics just by looking at the blueprint from the trunk. The difference between its prediction and the real metrics forms the confidence loss. Importantly, this loss only updates the confidence module, forcing it to become an expert critic without interfering with the main structure prediction network.
Training, Trends, and Frontiers
Beyond the main losses, several key strategies and design philosophies were essential for AlphaFold 3’s success, revealing a mature approach to building scientific AI.
Essential Training Strategies
-
Recycling: As in AlphaFold 2, the model’s own predictions can be “recycled” and fed back as input for another round of inference. This allows the model to iteratively correct its own mistakes—the computational equivalent of a student proofreading their own essay to find and fix errors.
-
Cropping: To manage the immense memory cost of training, complexes were randomly cropped. This included smart spatial cropping around interfaces, forcing the model to become an expert at modeling the all-important regions where molecules actually touch and interact.
Broader Impacts & ML Musings
The design choices in AlphaFold 3 reflect and inform the broader field of artificial intelligence.
-
The Rise of RAG in Science: The use of MSAs and templates is a form of Retrieval-Augmented Generation (RAG)[9]. AlphaFold pioneered this use of retrieved knowledge in science, and AF3 evolves the principle by showing that a more powerful core engine (the Pairformer and Diffusion Module) reduces the need for extensive retrieval, a lesson now being widely explored in large language models.
-
From Equivariance to Augmentation: AlphaFold 2 used complex, “equivariant”[10] modules like Invariant Point Attention—akin to building a sophisticated, expensive camera gimbal that’s immune to rotation. AlphaFold 3 takes a more pragmatic approach: data augmentation. It simply shows the model the same complex in thousands of random orientations. By seeing what stays the same, the model learns rotational invariance itself, rather than having it hard-coded. This is a significant shift towards simpler, more scalable, and often more robust components.
-
Pragmatic Engineering (Cross-Distillation): In a clever move, the training set included some structures predicted by the older AlphaFold-Multimer[11]. This taught AF3 to mimic its predecessor’s behavior for low-confidence regions (e.g., producing unstructured loops), providing users with a crucial and familiar visual indicator of uncertainty.
Conclusion
AlphaFold 3 is more than an incremental improvement; it’s a fundamental reimagining of what a structure prediction model can be. By shifting from a deterministic, protein-centric world to a universal, generative one, it has opened the door to tackling biological questions of unprecedented complexity. It moves us closer to a future where we can computationally model not just the individual parts of life’s machinery, but the complex, dynamic systems they form. This leap from predicting parts to modeling systems has tangible implications, promising to accelerate drug discovery, personalize medicine by understanding genetic mutations, and help design novel biomolecules. AlphaFold 2 solved a grand challenge; AlphaFold 3 provides a framework for exploring the entire biological universe that solution has unlocked.
Written on July 1, 2025
-
Jumper, J., Evans, R., Pritzel, A., et al. Highly accurate protein structure prediction with AlphaFold. Nature, 596(7873):583–589, 2021. ↩
-
Sengar, A. Deconstructing the Fold with AlphaFold 2. Blog Post, 2025. link ↩
-
Abramson, J., Adler, J., Dunger, J., et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature, 630(7930):493–500, 2024. ↩
-
While AlphaFold-Multimer later addressed protein-protein interactions, predicting how proteins interact with small drug molecules required a fundamental redesign. AlphaFold 3 was built specifically to capture these critical protein-drug interactions, providing researchers the clarity they need to quickly respond to new mutations. Evans, R., O’Neill, M., Pritzel, A., et al. (2022). Protein complex prediction with AlphaFold-Multimer. bioRxiv link. ↩
-
Google DeepMind Team and Isomorphic Labs. AlphaFold 3 predicts the structure and interactions of all of life’s molecules. Google Blog, May 8, 2024. link ↩
-
RDKit is a powerful open-source cheminformatics toolkit widely used in computational chemistry. In AlphaFold 3’s input pipeline, its main role is conformer generation: converting a 2D chemical graph into one or more plausible 3D structures. A single molecule can have multiple 3D shapes, or “conformers,” based on how its bonds are rotated, and this step provides a realistic initial 3D guess before AlphaFold 3 predicts how it will bind and interact within the larger biomolecular complex. ↩
-
Why 48 cycles? This number, like the specific channel dimensions used in the architecture, is a hyperparameter. It is not derived from a first-principles theory but is instead a value found through extensive experimentation and engineering trade-offs. The goal is to find a sweet spot: enough refinement cycles for the model to converge on an accurate structure, but not so many that the computational cost becomes prohibitive. This balance between predictive power and computational budget is a core aspect of building large-scale AI models. ↩
-
Further light readings on diffusion models: For a slightly more detailed and quick introduction to diffusion models in the context of alphafold, checkout the appendix section of this series [12]. For an accessible introduction to diffusion models in machine learning, see Lilian Weng’s blog[13]. For a slightly more math-inclined audience, check my blogpost on DDPMs [14]. ↩
-
Retrieval-Augmented Generation (RAG) is an AI technique where a generative model, instead of relying solely on its internal, pre-trained knowledge, can first retrieve relevant information from an external database at query time, and then use this retrieved context to generate a more accurate and informed response. ↩
-
An equivariant network is one where rotating the input causes the output features to rotate in the exact same way. ↩
-
Evans, R., O’Neill, M., Pritzel, A., et al. Protein complex prediction with AlphaFold-Multimer. Science, 378(6625):1169-1175, 2022. ↩
-
Sengar, A. Appendix (From Alphafold 2 to Boltz-2). Blog Post, 2025. link ↩
-
Weng, L. What are Diffusion Models? lilianweng.github.io (Blog Post), 2021. link ↩
